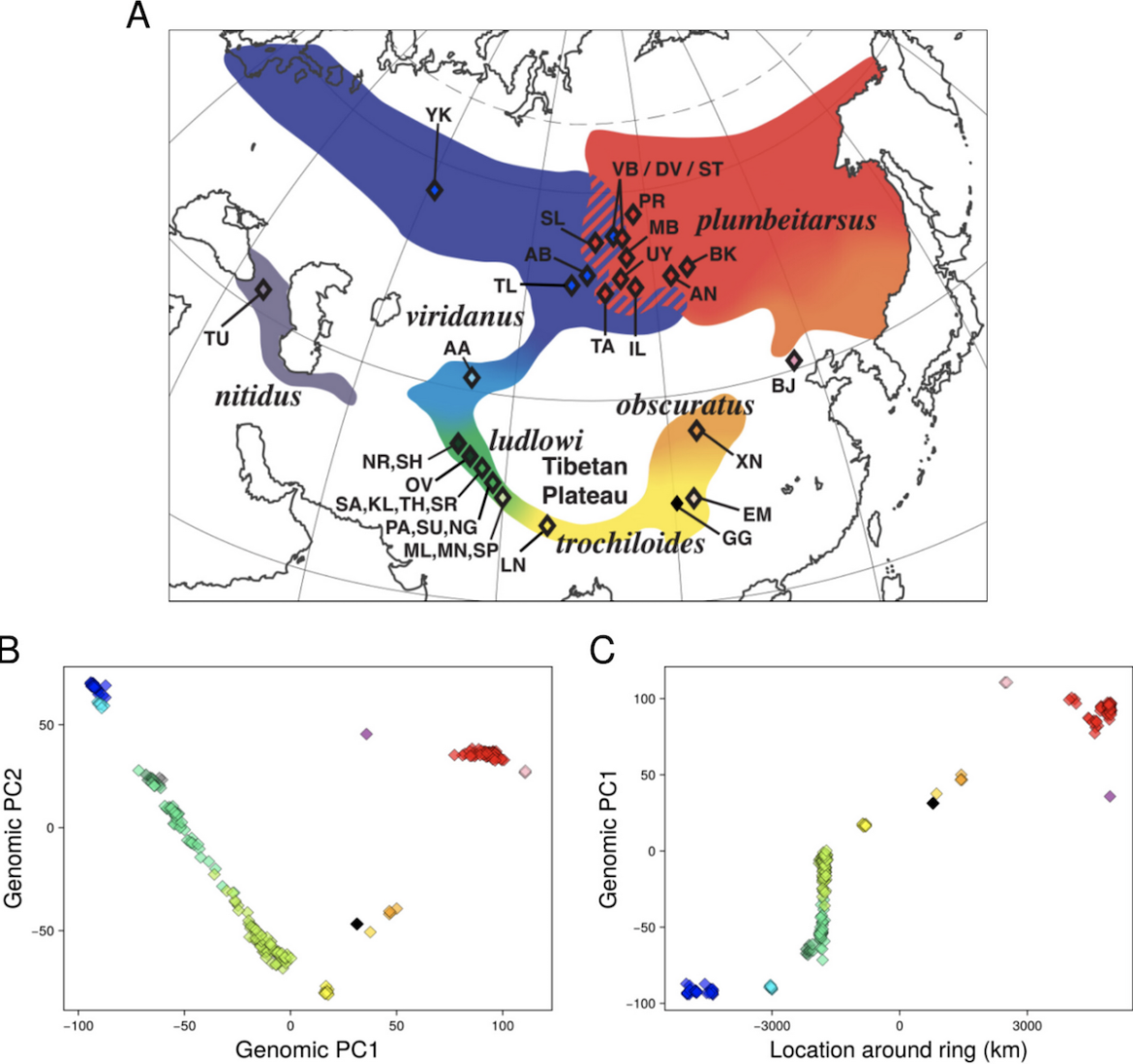
Figure 1. Greenish warblers show strong geographic structure, with west Siberian (P. t. viridanus) and east Siberian (P. t. plumbeitarsus) populations showing the most differentiation and populations to the south show stepwise progression in genetic signatures through subspecies ludlowi, trochiloides and obscuratus. (A) Map of sampling locations and subspecies ranges, (B) scatter plot of the first two principal components of genomic variation (PC1 captures 12.1% of the total variance, and PC2 6.4%), with each diamond representing one individual coloured according to the phenotype and map, and the black diamond showing the PCA coordinates of the reference genome. (C) The association of PC1 with location around the ring (measured from west Siberia down and around the ring to east Siberia). Note the western taxon nitidus is represented by two individuals in panel B (the two grey symbols near the upper left cluster of green symbols) but is not included in panel C because nitidus is outside of the main ring.
ABSTRACT
Haploblocks are regions of the genome that coalesce to an ancestor as a single unit. Differentiated haplotypes in these regions can result from the accumulation of mutational differences in low-recombination chromosomal regions, especially when selective sweeps occur within geographically structured populations. We introduce a method to identify large well-differentiated haploblock regions (LHBRs), based on the variance in standardised heterozygosity (ViSHet) of single nucleotide polymorphism (SNP) genotypes among individuals, calculated across a genomic region (500 SNPs in our case). We apply this method to the greenish warbler (Phylloscopus trochiloides) ring species, using a newly assembled reference genome and genotypes at more than 1 million SNPs among 257 individuals. Most chromosomes carry a single distinctive LHBR, containing 4–6 distinct haplotypes that are associated with geography, enabling detection of hybridisation events and transition zones between differentiated populations. LHBRs have exceptionally low within-haplotype nucleotide variation and moderately low between-haplotype nucleotide distance, suggesting their establishment through recurrent selective sweeps at varying geographic scales. Meiotic drive is potentially a powerful mechanism of producing such selective sweeps, and the LHBRs are likely to often represent centromeric regions where recombination is restricted. Links between populations enable introgression of favoured haplotypes and we identify one haploblock showing a highly discordant distribution compared to most of the genome, being present in two distantly separated geographic regions that are at similar latitudes in both east and central Asia. Our results set the stage for detailed studies of haploblocks, including their genomic location, gene content and contribution to reproductive isolation.